Florence Computational Biology Group
Escaping the ESKAPEs: integrated pipelines for new antibacterial drugs
General aim and project details:
With this proposal we aim to identify new compounds with promising antibacterial activity by three different perspectives and combining experimental and computational screenings/approaches.
Project duration: 25/04/2022 - 24/04/2025
Participating units:
RU1 and Project coordinator - IMPERI Francesco - Università di Roma Tre
RU2- FONDI Marco - Università degli Studi di FIRENZE
RU3 –BURONI Silvia- Università degli Studi di PAVIA
RU4 – SPERANDEO Paola - Università degli Studi di MILANO
RU5 – COLUCCIA Antonio - Università degli Studi di ROMA "La Sapienza"
Project code: 20208LLXEJ
Contributo MUR / costo totale: 853.920 Euro
Brief description of the project:
Contrasting antibiotic resistance is a priority of the public health agenda, and new antibacterial drugs are urgently needed. The characterization of novel cellular targets and the development of antimicrobials with novel mechanisms of action have become essential to tackle the spread of resistance and to control multidrug resistant bacterial infections.
The main goal of the present project is the identification, characterization, and pre-clinical assessment of novel antibacterial drugs active against multidrug resistant ESKAPE pathogens. The proposal has been organized into three WPs. WP1 is aimed at further characterizing two recently identified antibacterial agents that are mainly active against Gram-positive bacteria. In particular, their cellular targets, mechanisms of action and system-level effects on bacterial cells will be uncovered through the integration of different -omics techniques, and then validated by genetic, biochemical and structural analyses. The other WPs will be focused on the search for novel molecules active against Gram-negative bacteria that can interfere with cell envelope homeostasis or cell division. This aim will be pursued through two complementary approaches: (i) a structure-based virtual screening campaign against a panel of essential outer membrane biogenesis or divisome proteins (WP2), and (ii) the development of two high-throughput screening systems to screen a library of thousands of microbial extracts, followed by the activity-driven isolation of the active components from hit extracts (WP3). The newly identified antibacterial agents will be thoroughly characterized for potency, type and range of activity, cytotoxicity and efficacy in a simple animal model of infection. Moreover, -omics, computational and target-specific assays will be employed to decipher and/or confirm their mechanism of action. The proposal rests on solid foundations, due to the involvement of 5 research groups that share interest in antibacterials and drug discovery but with complementary scientific backgrounds and technical skills, and will take advantage of the participation as external collaborator of Naicons, an Italian biotech devoted to research on antibacterial drugs. Naicons discovered the two antibacterials analyzed in WP1, will provide the extracts library for the high-throughput screenings, and will collaborate to the isolation and purification of bioactive molecules. The expected deliverables of the project, in terms of both newly identified antibacterial compounds and information regarding their mode of action, would ultimately provide new pharmacological opportunities for the development of innovative therapeutic strategies to treat antibiotic resistant bacterial infections. Besides these direct deliverables, other major areas of project impact will be the industrial exploitation of results, high-quality scientific production, science dissemination and training of young scientists enrolled by the consortium.
What we do in the project:
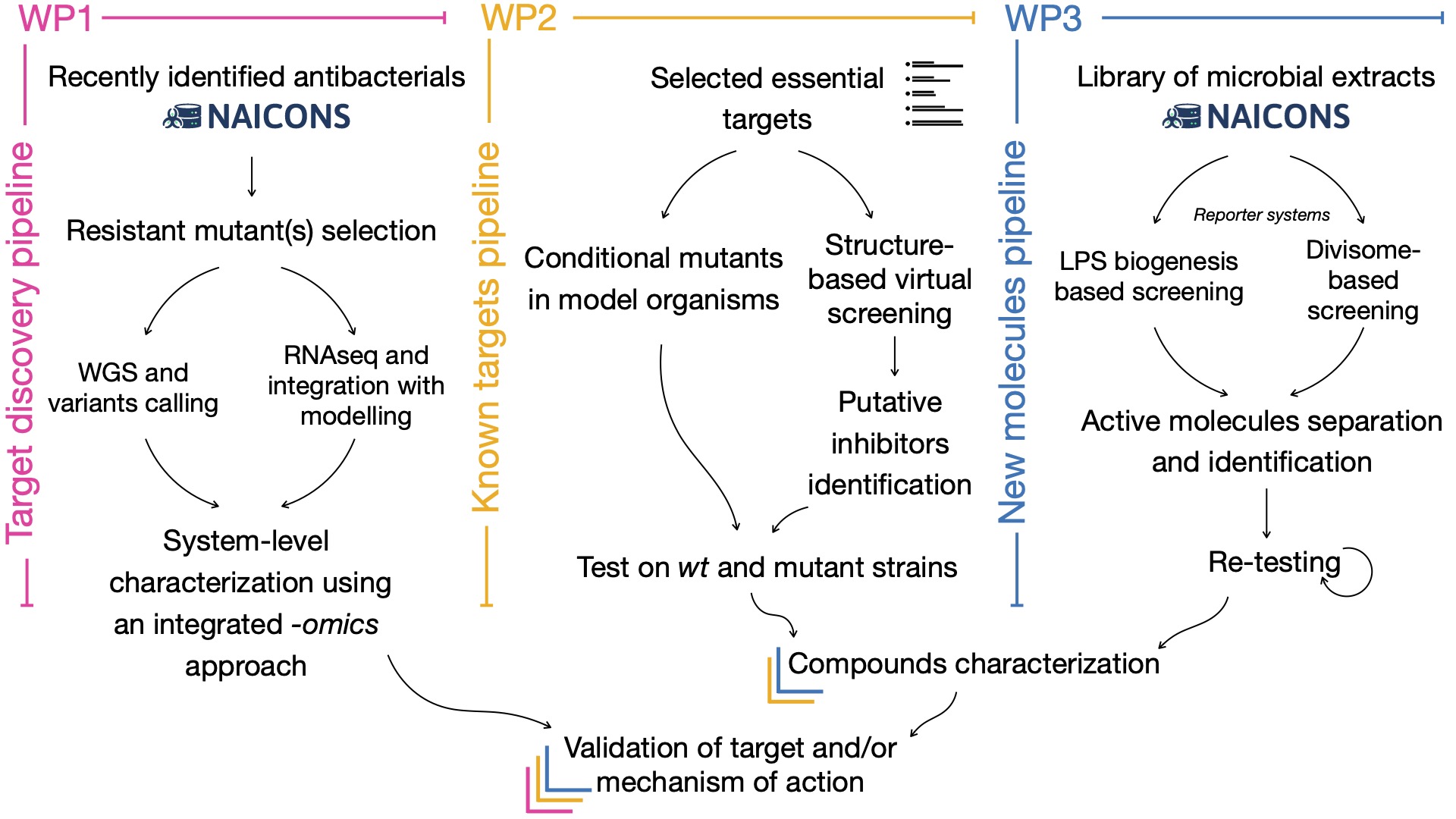
Main outputs of the project:
